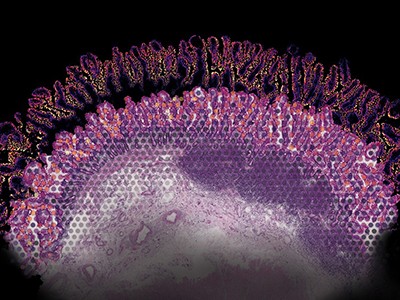
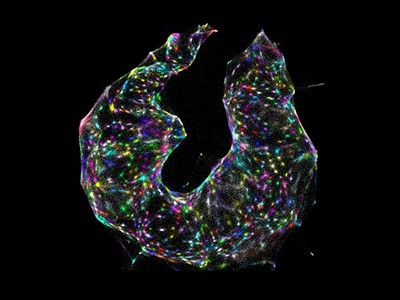
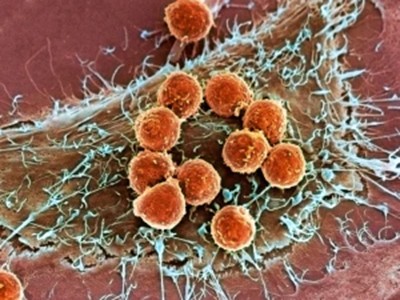
To individuals with most cancers, tumours can look like amorphous clumps of faulty cells, relentlessly targeted on unconstrained progress and invasion. However this doesn’t imply that they’re homogeneous. Cancerous cells have a broad spectrum of mutations, and growths include wholesome host cells, blood vessels and microscale fronts at which immune cells wage warfare with malignant tissue.
Till round a decade in the past, researchers had been ill-equipped to discover this tumour microenvironment. However the emergence of instruments that may spatially map massive numbers of biomolecules, resembling RNA and protein, has triggered one thing of a revolution. Certainly, researchers are more and more weaving these layers of data collectively to create wealthy ‘multiomic’ spatial maps that may classify various cell varieties and probe their actions all through a tumour.
Easy methods to make spatial maps of gene exercise — right down to the mobile stage
“We’re not simply speaking about tumour heterogeneity any extra — we are able to see it,” says Arutha Kulasinghe, a most cancers biologist on the College of Queensland in Brisbane, Australia. “We are able to see pockets of drug resistance, sensitivity and totally different biology instantly on the tissue.” The spatial elements that contribute to carcinogenesis and illness development are additionally more and more seen, revealing potential vulnerabilities within the course of. Such capabilities may rework most cancers analysis and pathology, making it potential to mannequin, interpret and maybe predict tumour biology with unprecedented sophistication.
However the boundaries to entry are excessive. There are numerous expertise platforms for spatial omics analyses, and the experiments will be expensive and complex. Even with information in hand, most cancers researchers can face a computational odyssey earlier than they’ll make sense of their outcomes. “All people desires what I prefer to name the ‘blender principle’ of multiomics, which is the place you throw all the info units collectively and it’ll inform you the reply as to what’s in them” says Elana Fertig, a bioinformatician at Johns Hopkins Medication in Baltimore, Maryland. “I’ve develop into much less and fewer satisfied that’s potential, as a result of everyone has a unique query that they wish to ask.”
Welcome to the neighbourhood
For greater than a decade, biologists have been learning tumour microenvironments by breaking tissue samples into particular person cells and characterizing their molecular contents. These single-cell omics applied sciences are pretty easy to make use of, not less than for RNA evaluation. Devices such because the Chromium from 10x Genomics in Pleasanton, California, can survey gene expression throughout thousands and thousands of particular person cells.
Some researchers, resembling most cancers genomicist Dan Landau on the New York Genome Heart in New York Metropolis, have even prolonged these instruments to carry out multiomic experiments — coupling transcription to different organic options, resembling genomic mutations or epigenetic alerts that instantly govern gene expression on the single-cell stage. “The imaginative and prescient is to attempt to begin understanding how these layers are speaking to at least one one other,” says Landau.
Such experiments can categorize cell varieties and reveal which organic processes these cells are engaged in — however they lack important context. “It was fairly clear early on that we miss numerous info by dissociating a tumour into single cells,” says Bernd Bodenmiller, a methods biologist on the College of Zurich in Switzerland and the Swiss Federal Institute of Expertise. For instance, the efficacy of immunotherapy in opposition to a given tumour relies upon not solely on which immune cells are current, but in addition the place they’re within the tumour.
In 2014, Bodenmiller helped to pioneer the spatial omics period when he and his colleagues mixed a laser ablation approach with mass spectrometry to detect and localize proteins labelled with varied metal-tagged antibodies (see Nature 567, 555–557; 2019). They referred to as the method imaging mass cytometry (IMC), and used it to quantify 32 proteins at subcellular decision in a breast-tumour specimen. Bodenmiller says that these early experiments demonstrated the significance of spatially localized communities of inter-communicating cells, now often known as ‘mobile neighbourhoods’, which might have been invisible utilizing dissociated single cells. “These had been the primary placing examples for me of how the spatial association of tumour cells — and the way they type communities with different cells — is actually strongly prognostic for affected person end result,” he says.
Most spatial experiments right now concentrate on the transcriptome, and there are quite a few industrial platforms accessible. Some are sequencing-based, such because the Visium platform from 10x Genomics, which builds on a technique developed in 2016 (see Nature 606, 1036–1038; 2022). Tissue slices are ready on a slide coated with an array of location-barcoded DNA strands. The RNA is then launched from the tissue, captured by these strands and transformed to DNA for sequencing; the barcode related to every sequence reveals the place it was on the slide.
Different strategies are imaging-based. For instance, the MERSCOPE platform from Vizgen in Cambridge, Massachusetts, is predicated on the approach MERFISH. First reported1 in 2015, the approach includes the serial labelling of tissue samples with fluorescently tagged probes that allow direct visualization, identification and quantification of transcripts in a specimen.
Starfish enterprise: discovering RNA patterns in single cells
The selection of platform includes trade-offs. “Usually, the imaging-based expertise can seize a bigger piece of tissue space, whereas with the sequencing-based [methods] you seize loads much less,” says Kai Tan, a analysis oncologist on the Youngsters’s Hospital of Philadelphia in Pennsylvania. Imaging-based strategies additionally have a tendency to supply superior spatial decision — right down to the mobile and even sub-cellular scale — however are extra labour-intensive and constrained, usually requiring customers to pick out which genes to probe somewhat than broadly interrogating the tissue RNA, and profiling a smaller fraction of the transcriptome than dissociated, single-cell strategies. Sequencing strategies can detect even sudden transcripts, albeit usually at decrease spatial decision. However “these two worlds are converging”, Landau notes.
For example, the Slide-tags technique affords an creative various, by which the address-defining barcodes for spatial transcriptomics are delivered instantly into the confines of the cell nucleus, offering subcellular decision2. These nuclei can then be remoted and analysed extra extensively with a spread of single-cell strategies.
Whatever the platform, spatial transcriptomics is unlocking thrilling alternatives for most cancers researchers. For instance, neurosurgeon Dieter Henrik Heiland on the College of Freiburg in Germany has used these strategies to tease aside the circumstances that foster the expansion and invasive behaviour of mind tumours, resembling glioblastoma — particularly, the influence of sure myeloid bone marrow cells on the exercise of immune system T cells. “We may determine outlined patterns, outlined architectures that we couldn’t do earlier than with every other applied sciences,” he says.
A multiplicity of maps
More and more, nonetheless, transcriptomics symbolize not everything of the spatial evaluation however one part thereof — a ‘baseline layer’, as Heiland places it. “Then, we predict what we are able to do on high.”
Usually, that’s spatial proteomics. Though all proteins are translated from messenger RNAs, not all mRNAs give rise to proteins. Kulasinghe says that in his expertise, spatial patterns of RNA and protein can differ by as much as 50% in a given pattern, such that transcription ranges may not reliably predict protein output. Proteins may also type complexes and bear chemical modifications that might be not possible to find out from transcriptomic information alone. Proteomic evaluation is due to this fact an important part in understanding tumour spatial biology. “Individuals are caught with proteins eternally,” says Garry Nolan, an immunologist at Stanford College in California.
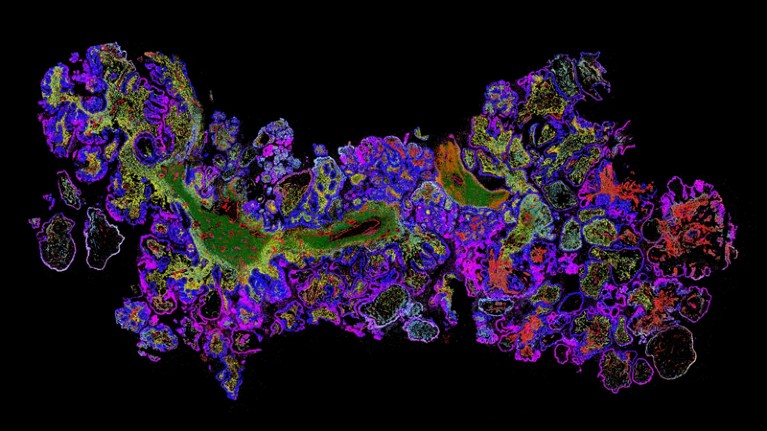
Information picture created utilizing the MERSCOPE platform of the genes in ovarian most cancers tissue.Credit score: Vizgen
At present’s spatial proteomics toolbox consists of strategies that may profile dozens and even lots of of proteins at a time. For instance, Nolan’s group developed3 the extensively used CODEX technique (now commercialized by Akoya Biosciences of Marlborough, Massachusetts, because the PhenoCycler system) in 2018. This method makes use of DNA-tagged antibodies for as much as 100 protein targets, that are sequentially detected with an enzymatic course of that particularly provides dye-labelled nucleotides to a subset of these DNA tags; these dyes are then cleaved off earlier than the subsequent imaging spherical. Equally, the GeoMx platform from NanoString in Seattle, Washington, permits researchers to picture RNA similtaneously a number of hundred proteins in the identical pattern.
Fertig and her group reported the mixed energy of spatial proteomics and transcriptomics in a research that explored the involvement of cells often known as fibroblasts within the development of premalignant pancreatic growths to most cancers4. “With the transcriptomics information, we had been capable of finding the fibroblasts and decide their influence on epithelial cells,” Fertig says. The tactic lacked the spatial decision to discriminate between cell varieties absolutely, however layering on IMC information revealed how some cancer-associated fibroblasts assist to ascertain a microenvironment that promotes malignant progress.
Maybe probably the most direct readout of what a cell is doing at any given second is the metabolome — the sugars, lipids, peptides and different biomolecules that act as inputs and outputs of organic processes. A number of teams are mapping the metabolome utilizing imaging mass spectrometry, by which a laser is scanned over a specifically ready pattern to generate spatially localized chemical signatures. “The fantastic thing about the expertise is you mainly get a very totally different image of your tissue than what you could have in your transcriptomic information,” says Heiland. In a single 2022 research, Heiland and his colleagues mixed this method with spatial transcriptomics and imaging mass cytometry to map out patterns of oxygen deprivation in glioblastoma. They discovered that hypoxic circumstances result in more-severe genomic disruption and irregular gene expression5.
Room for error
Nonetheless, spatial omics will be intimidating for newcomers. “All people desires to undertake spatial, but it surely’s overwhelming,” says Jasmine Plummer, a geneticist at St Jude Youngsters’s Analysis Hospital in Memphis, Tennessee. As head of the hospital’s Heart for Spatial Omics core facility, she advises customers to “take into consideration a particular query you wish to reply, not only a fishing expedition”, after which choose the strategy that gives the mandatory decision, multiplexing or different capabilities.
Some platforms permit customers to instantly survey a number of molecular classes directly. For instance, the NanoString GeoMx and CosMx devices can carry out each protein and gene-expression evaluation, and the Landau group collaborated with 10x Genomics to realize comparable analyses utilizing Visium6. However Bodenmiller cautions that in some circumstances, “you don’t get the optimum of every technique” with simultaneous analyses. For instance, the enzymatic digestion steps required to liberate RNA from tissue can harm proteins. However optimized workflows are rising to serially analyse the identical specimen utilizing a number of platforms, with the sample-preparation course of modified to reduce loss between steps. “I feel researchers have realized that you just want optimized expertise stacks,” says Kulasinghe.
Cell maps reveal contemporary particulars on how the immune system fights most cancers
Different teams carry out parallel analyses on consecutive skinny sections derived from the identical tumour pattern, then align and merge the ensuing information units. That is tougher than it sounds, nonetheless. “If you happen to go from one tissue part to the subsequent, you solely discover about 50–60% of the cells in each sections,” says Bodenmiller. Moreover, totally different experimental codecs can produce radically totally different information varieties, confounding integration. For instance, an IMC experiment yields an array of pixels denoting totally different proteins at subcellular decision, whereas sequencing-based transcriptomic experiments map ‘spots’ that always embody a number of cells. “Each information set needs to be handled by itself, and it’s important to work out how we are able to now combine these information by some type of similarity measurement,” says Heiland.
There may be additionally the basic problem of segmentation: precisely defining and classifying particular person cells within the spatial information. “If you happen to can not precisely section the boundary of the cell, then the whole lot downstream might be off,” says Tan. Totally different spatial platforms carry totally different challenges, and there aren’t any common options — Kulasinghe’s group has examined a number of algorithms for this goal and noticed inconsistent efficiency. As an answer, his group attracts boundaries based mostly on a ‘majority vote’ derived from a number of algorithms. Kulasinghe additionally emphasizes the significance of utilizing standard histology stains to truth test algorithmic analyses and set up ‘floor reality’ for a spatial research.
Above all, cautious planning is crucial. Spatial omics experiments are costly — Kulasinghe says {that a} single imaging-based transcriptomics assay can price practically US$10,000 — and might generate terabytes of information. “Getting pilot information on this realm is necessary,” says Plummer. “I don’t assume you wish to take an entire deep dive in till you’ve understood your information a bit of bit first.”
The ultimate frontier?
Fortuitously, the variety of core services is rising, giving researchers entry to skilled steering in addition to the technological capabilities wanted to carry out spatial analyses.
In parallel, worldwide and cross-institutional analysis efforts are leveraging single-cell — and, more and more, spatial — multiomic evaluation at unprecedented scale, together with the US Nationwide Institutes of Well being-backed Human Tumour Atlas Community and world consortium the Human Cell Atlas. These efforts are creating and optimizing analytical pipelines and instruments, and, extra importantly, producing huge collections of reference information for wholesome and diseased tissues that the scientific neighborhood can use to interpret future experiments. “With out these massive initiatives, we’d actually not be on the state of expertise and risk the place we are actually,” says Bodenmiller.
NatureTech hub
In the meantime, some spatial-omics pioneers wish to new horizons. Bodenmiller’s group reported a substitute for IMC by which antibodies are labelled with isotopic tags that may be detected and distinguished by X-ray imaging somewhat than mass spectrometry, permitting fast mapping of many proteins directly all through the specimen7. He says that the strategy could possibly be a superb match for 3D imaging, and is quick as a result of it avoids the gradual scanning course of that’s typical of IMC. First, nonetheless, the group should work out logistical challenges, resembling the right way to effectively ship antibodies into the inside of intact tissues.
The approaching deluge of spatial information may even present a treasure trove for researchers trying to apply deep-learning strategies to most cancers. This consists of ‘digital pathology’ methods, by which artificial-intelligence algorithms are educated to correlate options on standard pathology slides with molecular indicators which can be related to tumour id, prognosis and susceptibility to remedy. Firms are already getting into this area with assays that information drug choice based mostly on spatial information, and Kulasinghe sees alternatives to evaluate immune exercise in a tumour or predict the probability of metastasis with out the necessity for spatial assays within the clinic. “This can provide us deeper insights into the tumour microenvironments that in the end affiliate with medical endpoints,” he says.
For his half, Nolan predicts a post-data world, by which the analysis precedence shifts from producing molecular maps to utilizing them to coach AI fashions that reveal hidden vulnerabilities. “We’re going to have the ability to create a digital tissue that appears similar to colon most cancers,” he says. “Then you can begin to alter the parameters, and say: ‘OK, how do I cease the next construction from forming?’”